A user-friendly multi-omics data harmonisation R pipeline
Published in f1000, 2021
Recommended citation: Tyrone Chen, Al J Abadi, Kim-Anh Lê Cao, Sonika Tyagi, "multiomics: A user-friendly multi-omics data harmonisation R pipeline [version 1; peer review: awaiting peer review]" F1000Research July 2021, 10:538. DOI: https://doi.org/10.12688/f1000research.53453.1 https://doi.org/10.12688/f1000research.53453.1
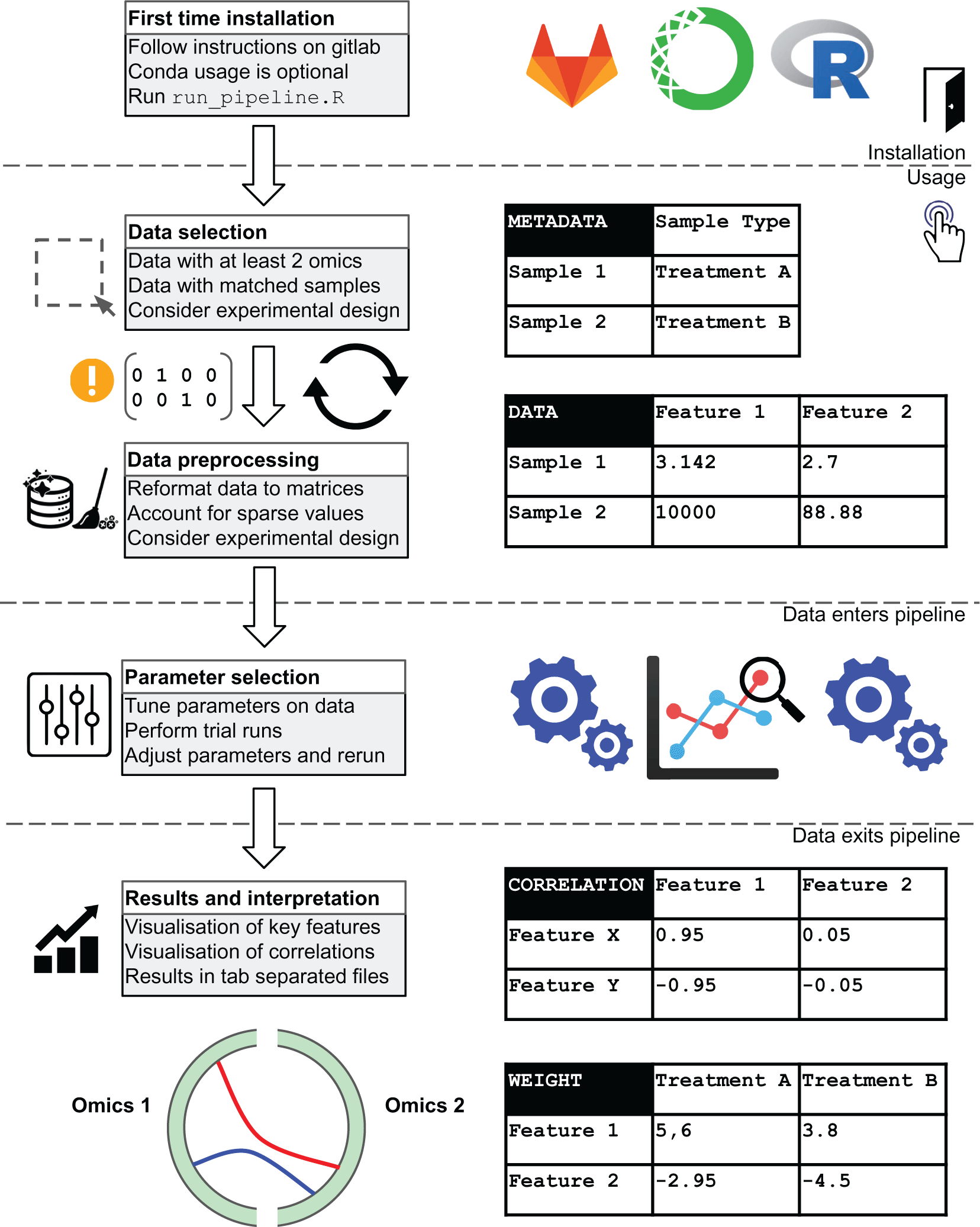
We present our R multiomics pipeline as an easy to use and flexible pipeline that takes unrefined multi-omics data as input, sample information and user-specified parameters to generate a list of output plots and data tables for quality control and downstream analysis.
Plain text citation:
Tyrone Chen, Al J Abadi, Kim-Anh Lê Cao, Sonika Tyagi, "multiomics: A user-friendly multi-omics data harmonisation R pipeline [version 1; peer review: awaiting peer review]" F1000Research<\i> July 2021, 10:538. DOI: https://doi.org/10.12688/f1000research.53453.1
Bibtex citation:
@article{chen2021multiomics,
title={multiomics: A user-friendly multi-omics data harmonisation R pipeline},
author={Chen, Tyrone and Abadi, Al J and L{\^e} Cao, Kim-Anh and Tyagi, Sonika},
journal={F1000Research},
volume={10},
number={538},
pages={538},
year={2021},
publisher={F1000 Research Limited}
}
